So good to hear so much form you, Faris, please keep going! I would love to hear from the rest of you as well though, so I can keep updated on the research going on. I’m hoping I’ll be able to start updating regularly soon as well, as I get boards out of the way!
Right so some plaques were found Today or…
Right, so some plaques were found! Today or yesterday even which is good news. About time I got some of them bad boys. On Monday or during the week these plaques could be potentially isolated and analysed which is exciting. At the same time I am going to try and produce some plaques from making more plates which allows analysis of many as poss. This could be a good week so Ill update as much as poss.
Today no plaques were found which was pretty…
Today no plaques were found which was pretty disappointing but slight change of the method is to be undergone which will develop a method to produce a good plaque assay. Although no plaques were formed, Its just the beginning of the journey for me and its pretty cool….
Right so today’s been a pretty good day…
Right so today’s been a pretty good day. My Broths were looking pretty good which was a good start. Managed to make some soft agar in tubes which was pretty cool. My nutrient agar was ready and I managed to mix my e.coli broth with it to make my pour plates.After a lovely big mac meal with free cheeseburger I isolated my phages using a centrifuge and a syringe containing 0.22 micron filter. I then mixed these with the soft agar. The soft agar containing the phages was placed on the top of the pour plates. This was then incubated at 37 degrees Celsius so the phages can interact with the e.coli in the nutrient agar. So to summarise the theory: The plates contain a bottom layer containing nutrient agar and e.coli where the top layer contains soft agar with the phages so hopefully the phages will interact with the e.coli overnight. I know you Biomed know this already so soz. Although I might not get the results first time, I now have confidence in doing plaque assays and feel this project could be a pretty sick one.
A question I would like to pose to…
A question I would like to pose to everyone on this blog, as stupid as it may seem. Is there a method to improve the resolution of an oil immersed, KÖHLER illuminated light microscope image?
I am very happy to have finally been…
I am very happy to have finally been able to gain access to this blog, after a week of being in the laboratory. Although I was expecting my experience to be productive, it has so far been more productive and far more useful than I previously expected. My group is currently working on assessing the presence of antibiotic resistant bacteria (specifically gram negative) in “ready to eat” food from sources within and outside of the UK. In our first week we were given the opportunity to produce our own practical protocol and method of logging results which I have found hugely useful. In addition I have managed to re-cap over most of the micro biology practical skills that I struggle with. As a result whilst I still find some methods tricky I find them far less tricky as a result of my experience so far.
Today was the first practical day which involved…
Today was the first practical day which involved making agar, making an e.coli broth which finger crossed will be ok and 6 tubes of nutrient broth containing a small volume of Brayford pool water. These have all been left to incubate and tomorrow I will be starting the soft agar and isolating phages if all goes to plan….
Just a quick update to say hello again…
Just a quick update to say hello again, and hope that you manage to get online and blogging now! Following the session this afternoon on forms, have a go at filling them in as best as you can, and then bring them into the lab to show us what you’ve managed. Use this summer as a way of finding out as much as you can about the behind the scenes parts of scientific research, as this is the area that will put you way ahead of the game in September!
As for my research, I’ve only just managed to sub on my E. coli strains today that I grew on Thursday of last week, so it’s been a slow start!
Howdy all For those that don’t know I’m…
Howdy all,
For those that don’t know, I’m always floating around the lab under some guise. Currently I’m finishing up some written work, shortly I will be back to my PhD research which will involve some PCRs that I’ve designed and cloning the products into E.coli. I will also be getting some more sequencing work done, and then after taking a bit of a break (I need a holiday!) will be looking at some protein expression.
Since you can see all the previous posts from last year, I was responsible for supervising the PFGE project on P.acnes and the videoblogging which was really successful and is being presented at this years Student As Producer conference.
Get blogging!!! Honestly, this is a good platform for you to think/reflect/discuss and log what you’ve done. It will be worth it.
Get involved 😀
Hi my name is Faris and I have…
Hi my name is Faris and I have just finished second year of the Forensic Science degree. I have received some funding from the University to carry out a project based on the analysis of antibiotic resistance genes present in bacteriophages. Im pretty excited and should be starting this week and would appreciate help from anyone in the lab and will try and return the favour.
Today I made some plates and restreaked some…
Today I made some plates and restreaked some Paracoccus strains. Let’s see if they grow…
Welcome to all our new students and welcome…
Welcome to all our new students, and welcome back to those who contributed last year. Do please introduce yourself and let us know what you’re working on here, and keep us updated with your progress.
This year I’ll carry on with some of my plasmid stability work, but I’m also working on background fibre research, with the hope of getting a couple of papers out – watch this space!
Last day in the lab today Quite unexpectedly…
Last day in the lab today 🙁
Quite unexpectedly the spread plates yesterday grew very well, over 100 colonies on each plate. I carried on to replica plate the ones with close to 100 colonies and incubated them over night at 37C. These also grew very well, i documented these down but i am not sure of what these results mean (if anything) as T0 did not yield enough growth to replica plate. Because it was just a plate count day i did pcr, using my hair, just as the results from before im a boy!?!
As i expected pALA1029 pOG04 DRW and pOG4003…
As i expected, pALA1029, pOG04 DRW and pOG4003 have once again shown no growth, almost in sympathy the serial dilution i performed yesterday yielded poor results. The spread plates only showed colony growths of 1-14, as i was looking for colonies around 100, i decided that it was not worth replica plating. I have still carried out a serial dilution from the overnight broth cultures and spread plated them onto LB agar plates and left them to incubate at 37C overnight. I do not expect any growth based on todays results but one can hope!
Another frustrating day in the lab Grew pNC0412B…
Another frustrating day in the lab!!
Grew pNC0412B to an OD of 1 then performed a serial dilution (A,B,C,D -1to-5.5), I innoculated a fresh 10ml LB broth with 100 microlitres from dilution -4 and placed into the 37C shaker overnight. I then spread plated 100 microlitres from dilution -5 onto a fresh LB agar plate, I also spread plated 100 microlitres from dilution -5.5 onto a fresh LB agar plate, then placed these into the 37C incubator overnight. I would have also done this for pOG04 NJC but it refused to grow, after 6 hours in the 37C shaker only an OD of 0.01 was achieved, i abandoned this set of broths. The plates i hoped to grow for today containing the plasmids pALA1029, pOG4003 and pOG04 DRW also showed no growth. I have tried again today so hopefully tomorrow will be a better day. I seriously think C2110 is the work of the devil!
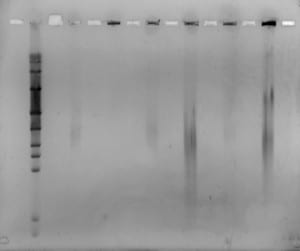
Hello Today was our last day in the…
Hello!
Today was our last day in the lab! 🙁 We filmed our latest and final videoblog, removed the gel, stained and viewed it. We saw some separation from all but one of the plugs, however separation was very blurred. We decided to run the same samples again, but for longer and with a higher switch interval. We set up and ran a new gel for 18 hours, at 6V/cm, with a 1-30 second switch interval, a 1.4% gel with modified buffer flow.
Mike will remove the gel tomorrow, stain it and view for us. Hopefully we will see better separation, from the samples containing enough DNA.
The last 10 weeks have been amazing – i have learnt so much since i started and it’s going to be really odd to not be in the lab everyday. I have really enjoyed working through a project, having to work on problems and figure out solutions. Then getting to see the good and bad results. I am very impressed with myself, as i set out to blog everyday i was in the lab with what i have done, the good and the bad! and i have actually managed it! 🙂 I wasn’t sure about whether i would get on with videoblogging at the beginning, but i have really enjoyed filming them every week – although most the content was never included as we ALWAYS got distracted! I really hope those who have watched them have found them as interesting and enjoyable as we did.
It has been a very rewarding experience, and i am so grateful to everyone who has made the last 10 weeks, the best summer – you all know who you are! And of course Mike, who has put up with us and our random chatter, better than i ever imagined he would. It has been an absolute pleasure to work along side him, and i hope i get the chance to do it again.
Today may be Friday but because of the…
Today may be Friday but because of the bank holiday monday, i am treating today like a monday!!
I am growing two batches of C2110 up to an OD of 0.35. Then i will perform cell competancy and transformation. Normally i would put the spread plates in the 37C incubator overnight but because of the bank holiday i am placing the plates in the 25C until tuesday. Hopefully there will be some growth. If all goes well Tuesday i will be working with pNC0412B, pOG04NJC, pALA1029 and pOG4003.
Hey All Removed plugs from SpeI enzyme buffer…
Hey All,
Removed plugs from SpeI enzyme, buffer and BSA, and washed in 1x wash buffer for 30mins. We removed this, then incubated them in 0.5x TAE buffer for an hour, while we prepared a 1.4% agarose gel. We then loaded 1/2 of each plug strain into individual wells, as well as the low range PFG marker, sealed with agarose and ran at 6V/cm, for 15 hours, with a 1-12 sec switch interval, and modified buffer flow.
Tomorrow, we will remove the gel, stain it and view it. We will also be filming our last videoblog! 🙁 as it will be the last day of our project – 10 weeks has gone by sooo fast! We may also be running another gel, depending on the results of the first. See you all then 🙂
Had growth from two of my plasmids pOG04…
Had growth from two of my plasmids, pOG04 DRW and pNC0412B but nothing from pOG04 NJC. I used an original sample of pNC0412B and so i am very happy that there is growth, this means that there is probably a fault with my plasmid prep. At least i have something to work with. The pOG04 NJC is a known temperemental plasmid so no growth is not wholly unexpected, just frustrating. Today just counted my T end plates and recorded the data, the rest of the day is spent making up agars and broths ready for next week as it is the last week in the lab and i would like to be prepared!
Hello Today we removed the plugs from proteinase…
Hello,
Today we removed the plugs from proteinase K, and washed them in 1 x wash buffer 4 times, for an hour each. We then incubated them in 0.1 x wash buffer for an hour, then in 1 x SpeI buffer to saturate for an hour. We then removed and added fresh SpeI buffer, SpeI enzyme and BSA and incubated overnight at 37C. We also made up 500ml of 0.5x TAE buffer and autoclaved it for tomorrow.
Unfortunately, we lost a few plugs along the way, due to instability. Tomorrow we will be incubating the plugs in 1 x wash buffer, then 0.5x TAE, before running the new plugs.